Note
Go to the end to download the full example code.
DA with Random Initial Conditions on Soil Layers#
This notebook demonstrates how to create an ensemble of models with varying initial conditions for different soil layers. By introducing randomness into the initial conditions, we can better simulate the natural variability in soil properties and improve the robustness of data assimilation (DA) processes.
Estimated time to run the notebook = 2min
import os
import numpy as np
import pyvista as pv
from pyCATHY.DA.perturbate import perturbate_parm
from pyCATHY.DA import perturbate
from pyCATHY.DA.cathy_DA import DA
from pyCATHY.plotters import cathy_plots as CTp
nlay = 6
scenario = {
'per_name':['ic'],
# 'per_type': [[None]*nlay],
'per_type': [['multiplicative']*nlay],
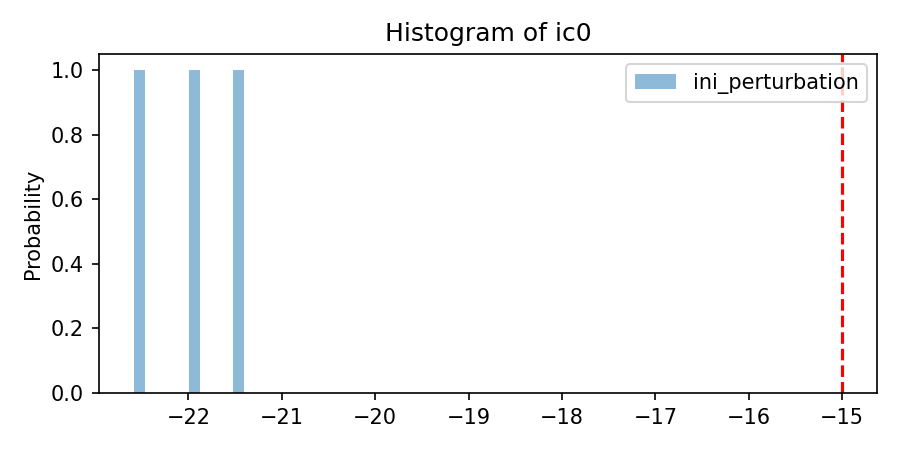
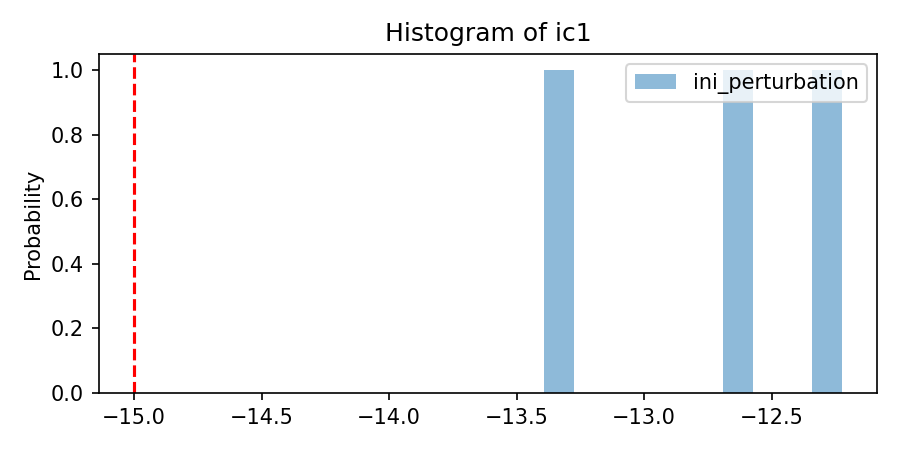
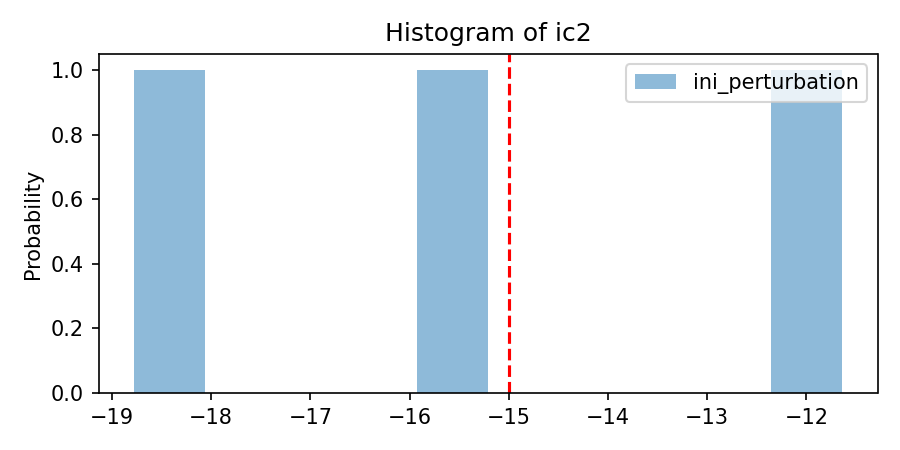
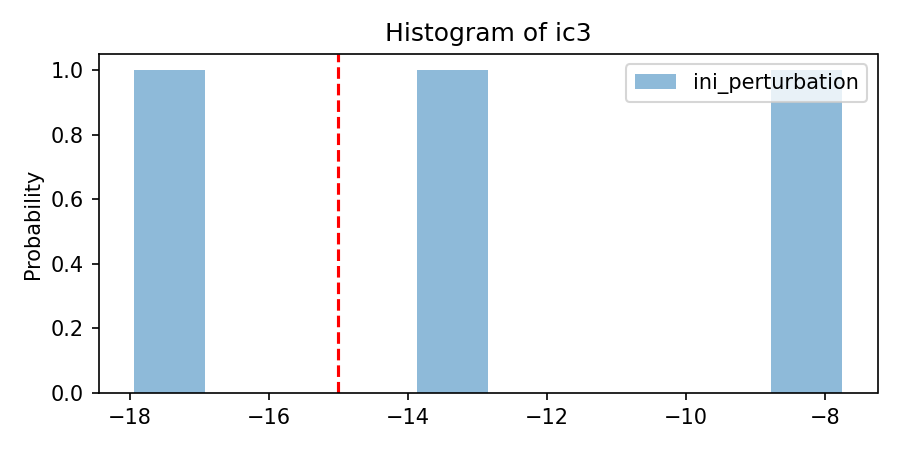
'per_nom':[[-15]*nlay],
# 'per_mean':[[-15]*nlay],
'per_mean':[[1]*nlay],
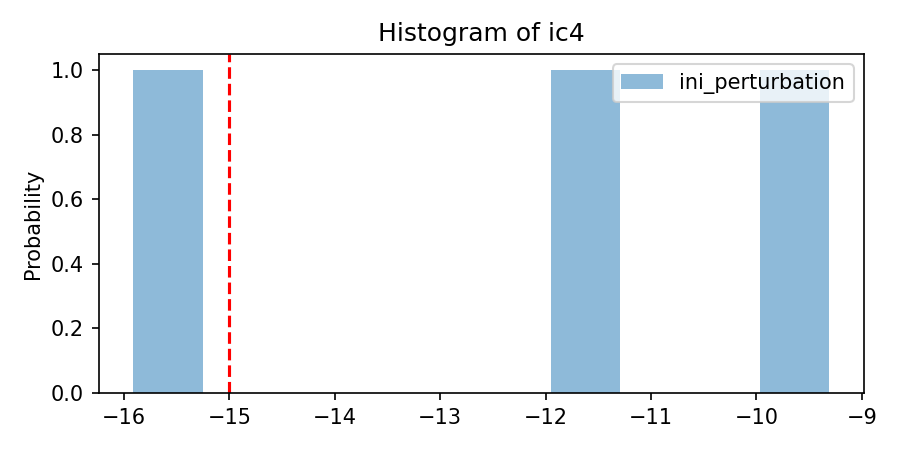
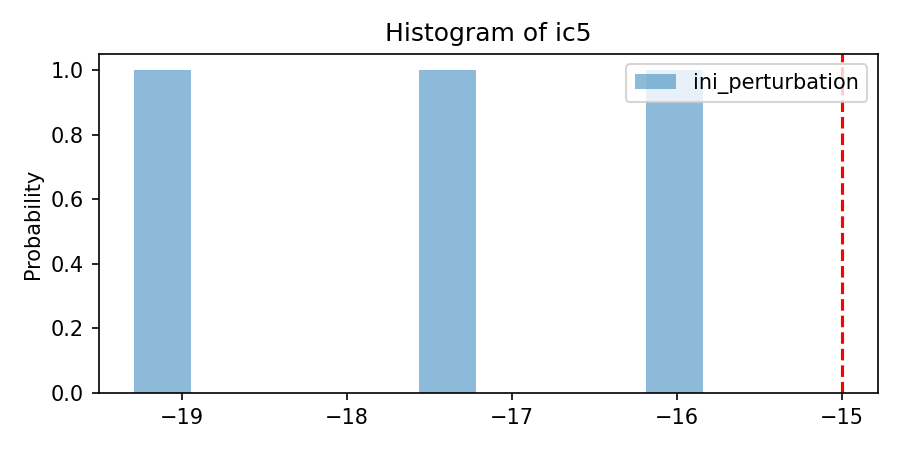
'per_sigma': [[3.75]*nlay],
'per_bounds': [[None]*nlay],
'sampling_type': [['normal']*nlay],
'transf_type':[[None]*nlay],
'listUpdateParm': ['St. var.'],
'pert_control_name': ['layers']*nlay
}
simuWithDA = DA(
dirName='./',
prj_name= 'DA_with_non_uniform_ic',
notebook=True,
)
# linear z depth
# ---------------
zb = np.linspace(0, 2, nlay)
nstr = len(zb)
zr = list(np.ones(len(zb))/nstr)
simuWithDA.update_prepo_inputs(
nstr=nstr,
zratio=zr,
base=max(zb),
)
🏁 Initiate CATHY object
🔄 Update hap.in file
🔄 Update dem_parameters file
🔄 Update dem_parameters file
simuWithDA.update_dem_parameters()
simuWithDA.update_veg_map()
simuWithDA.update_soil()
🔄 Update dem_parameters file
────────────────────────── ⚠ warning messages above ⚠ ──────────────────────────
The parm dictionnary is empty
Falling back to defaults to update CATHYH
This can have consequences !!
────────────────────────────────────────────────────────────────────────────────
🔄 Update parm file
🔄 Update soil
homogeneous soil
simuWithDA.NENS = 3
list_pert = perturbate.perturbate(
simuWithDA,
scenario,
simuWithDA.NENS,
)
var_per_dict_stacked = {}
for dp in list_pert:
var_per_dict_stacked = perturbate_parm(
var_per_dict_stacked,
parm=dp,
type_parm = dp['type_parm'], # can also be VAN GENUCHTEN PARAMETERS
mean = dp['mean'],
sd = dp['sd'],
sampling_type = dp['sampling_type'],
ensemble_size = dp['ensemble_size'], # size of the ensemble
per_type= dp['per_type'],
nlayers = nlay,
savefig= os.path.join(simuWithDA.workdir,
simuWithDA.project_name,
simuWithDA.project_name + dp['savefig'])
)
/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/scipy/stats/_qmc.py:993: UserWarning: The balance properties of Sobol' points require n to be a power of 2.
sample = self._random(n, workers=workers)
This in normally directly called when using run_DA_sequential()
simuWithDA.ens_valid = list(np.arange(0, simuWithDA.NENS))
simuWithDA._create_subfolders_ensemble()
simuWithDA.apply_ensemble_updates(var_per_dict_stacked,
var_per_dict_stacked.keys(),
cycle_nb=0
)
Traceback (most recent call last):
File "/home/runner/work/pycathy_wrapper/pycathy_wrapper/examples/DA/plot_1_DA_NoUni_ic.py", line 97, in <module>
simuWithDA.apply_ensemble_updates(var_per_dict_stacked,
File "/home/runner/work/pycathy_wrapper/pycathy_wrapper/pyCATHY/DA/cathy_DA.py", line 2151, in apply_ensemble_updates
ic_nodes = self.map_prop_2mesh_markers('ic',
File "/home/runner/work/pycathy_wrapper/pycathy_wrapper/pyCATHY/cathy_tools.py", line 3699, in map_prop_2mesh_markers
mt.add_markers_zone3d_2_mesh(
File "/home/runner/work/pycathy_wrapper/pycathy_wrapper/pyCATHY/meshtools.py", line 932, in add_markers_zone3d_2_mesh
_find_nearest_point2DEM(
File "/home/runner/work/pycathy_wrapper/pycathy_wrapper/pyCATHY/meshtools.py", line 874, in _find_nearest_point2DEM
mesh_pv_attributes.save(saveMeshPath,binary=False)
File "/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/pyvista/_deprecate_positional_args.py", line 248, in inner_f
return f(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/pyvista/core/dataobject.py", line 213, in save
raise FileNotFoundError(msg)
FileNotFoundError: Parent directory does not exist: /home/runner/work/pycathy_wrapper/pycathy_wrapper/examples/DA/DA_with_non_uniform_ic/DA_Ensemble/cathy_1/vtk
pl = pv.Plotter(shape=(1,2))
for i, ensi in enumerate([1,3]):
DApath = f'DA_Ensemble/cathy_{ensi}/vtk/'
path = os.path.join(simuWithDA.workdir,
simuWithDA.project_name,
DApath,
simuWithDA.project_name + '.vtk'
)
pl.subplot(0,i)
CTp.show_vtk(path,
'ic_nodes',
ax=pl,
clim = [-25,-5],
#show_scalar_bar=True,
)
_ = pl.add_legend('')
pl.add_title(f'Ensemble nb:{ensi}')
pl.show()
Total running time of the script: (0 minutes 18.064 seconds)