Note
Go to the end to download the full example code.
Read SMC sensors observations to assimilate#
The notebook illustrate how to read SMC sensors dataset to be prepare for DA
Estimated time to run the notebook = 2min
import numpy as np
from pyCATHY.DA.cathy_DA import DA
import pandas as pd
import matplotlib.pyplot as plt
from pyCATHY.DA.cathy_DA import DA, dictObs_2pd
from pyCATHY.DA.observations import read_observations, prepare_observations, make_data_cov
from pathlib import Path
import pyvista as pv
simuWithDA = DA(
dirName='./DA_with_swc',
prj_name='import_SMC',
notebook=True,
)
🏁 Initiate CATHY object
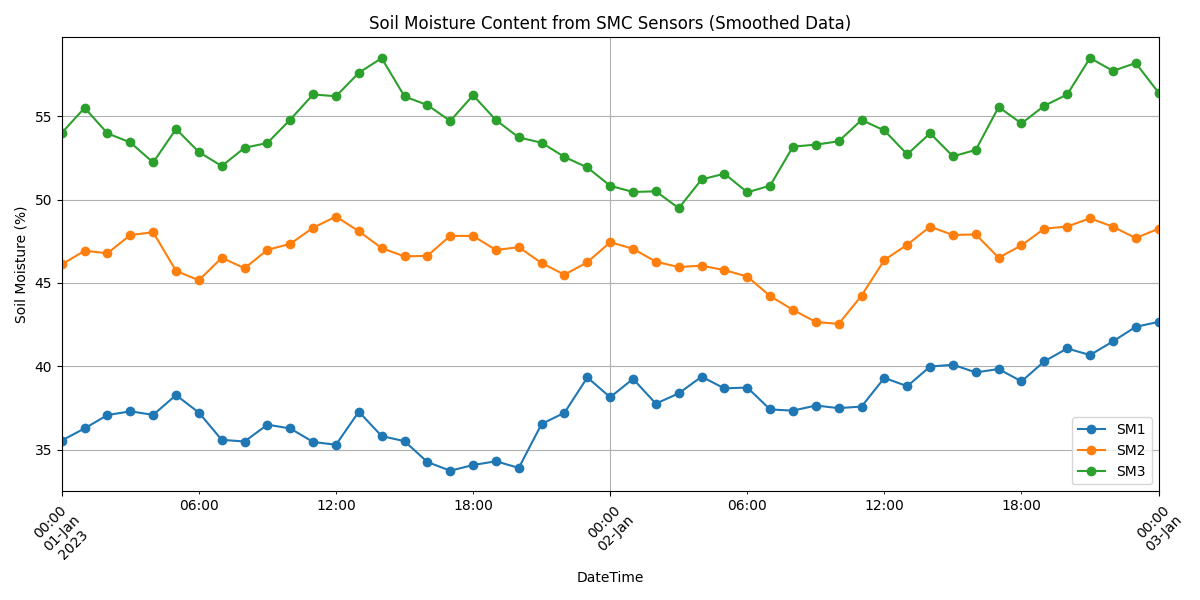
date_range = pd.date_range(start='2023-01-01', end='2023-01-03', freq='H')
# Create smoother soil moisture data using cumulative sum of random changes
sm_data = {f'SM{i}': np.cumsum(np.random.normal(0, 1, len(date_range))) + 25 + 10*i for i in range(1, 4)}
# Create a dataframe with smoother data
sm_smooth = pd.DataFrame(sm_data, index=date_range)
# Plot the smoother soil moisture data
sm_smooth.plot(figsize=(12, 6), marker='o')
plt.xlabel('DateTime')
plt.ylabel('Soil Moisture (%)')
plt.title('Soil Moisture Content from SMC Sensors (Smoothed Data)')
plt.legend()
plt.xticks(rotation=45)
plt.grid(True)
plt.tight_layout()
plt.show()
sm_smooth['Seconds'] = (sm_smooth.index - sm_smooth.index[0]).total_seconds()
simuWithDA.create_mesh_vtk()
SMC_XY = [5,5]
SMC_depths = [0.05,0.25,0.75] # SMC depths
# find the altitudes of the nodes at the mesh position x, y = (0.05,0.25)
_ , closest = simuWithDA.find_nearest_node(SMC_XY)
nodes_SMC = []
closestPos = []
for d in SMC_depths:
SMC_XYZi = [5,5,closest[0][2]-d]
nodeId, closest = simuWithDA.find_nearest_node(SMC_XYZi)
nodes_SMC.append(nodeId)
closestPos.append(closest)
nodes_SMC = np.hstack(nodes_SMC)
SMC_XYZ = np.vstack(closestPos)
pl = pv.Plotter(notebook=True)
mesh = pv.read(
Path(simuWithDA.workdir) /
simuWithDA.project_name /
f'vtk/{simuWithDA.project_name}.vtk'
)
pl.add_mesh(mesh,
opacity=0.7
)
pl.add_points(SMC_XYZ,
color='red'
)
pl.show_grid()
pl.show()
🍳 gfortran compilation
👟 Run preprocessor
🔄 Update parm file
🔄 Update hap.in file
🔄 Update dem_parameters file
🔄 Update dem_parameters file
🛠 Recompile src files [3s]
🍳 gfortran compilation [8s]
b''
👟 Run processor
/home/z0272571a@CAMPUS.CSIC.ES/miniconda3/envs/myenv/lib/python3.10/site-packages/pyvista/jupyter/notebook.py:36: UserWarning: Failed to use notebook backend:
cannot import name 'vtk' from 'trame.widgets' (/home/z0272571a@CAMPUS.CSIC.ES/miniconda3/envs/myenv/lib/python3.10/site-packages/trame/widgets/__init__.py)
Falling back to a static output.
warnings.warn(
<PIL.Image.Image image mode=RGB size=2048x1536 at 0x7F72277E8AF0>
abs_data_err = 1e-1 # constant error does not vary with time
dict_obs = {} # initiate the dictionnary
for i in range(len(sm_smooth.columns)-1):
for j, assimilation_time_sec in enumerate(sm_smooth['Seconds']):
dict_obs = read_observations(
dict_obs,
obs_2_add=sm_smooth[sm_smooth.columns[i]].iloc[j],
tA=assimilation_time_sec,
mesh_nodes = nodes_SMC[i],
data_type='swc',
data_err=abs_data_err,
colname=' m³/m³ Water Content',
datetime=sm_smooth.index[j]
)
data_measure_df = dictObs_2pd(dict_obs)
data_measure_df
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
already existing assimilation time
By default, there is no correlation between sensors Therefore, the covariance matrices are diagonal with the error values on the diagonals
_,_, stacked_data_cov = make_data_cov(
simuWithDA,
dict_obs,
list_assimilated_obs = 'swc',
)
print(np.shape(stacked_data_cov))
simuWithDA.stacked_data_cov = stacked_data_cov
(49, 9, 9)
import pickle
with open('obs_prepared_SMC.pkl', 'wb') as fp:
pickle.dump(dict_obs, fp)
Total running time of the script: (0 minutes 9.370 seconds)