Note
Go to the end to download the full example code.
Update with spatially and temporally distributed atmospheric boundary conditions (bc)#
This tutorial demonstrates how to update atmospheric boundary conditions (bc) using spatially and temporally distributed data in a hydrological model.
Reference: Weill, S., et al. « Coupling Water Flow and Solute Transport into a Physically-Based Surface–Subsurface Hydrological Model ». Advances in Water Resources, vol. 34, no 1, janvier 2011, p. 128‑36. DOI.org (Crossref), https://doi.org/10.1016/j.advwatres.2010.10.001.
This example uses the pyCATHY wrapper for the CATHY model to reproduce results from the Weill et al. dataset. The notebook is interactive and can be executed in sections to observe the intermediate results. It can also be shared for collaborative work without any installation required.
Estimated time to run the notebook = 5 minutes
# Import necessary libraries
import os
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pyvista as pv
# Import pyCATHY modules for handling mesh, inputs, and outputs
import pyCATHY.meshtools as mt
from pyCATHY import cathy_tools
from pyCATHY.importers import cathy_inputs as in_CT
from pyCATHY.importers import cathy_outputs as out_CT
from pyCATHY.plotters import cathy_plots as cplt
Define the project directory and model name. This example uses ‘atmbc_spatially_from_weill’.
path2prj = "../SSHydro/" # Replace with your local project path
simu = cathy_tools.CATHY(dirName=path2prj, prj_name="atmbc_spatially_from_weill_withnodata")
figpath = "../results/DA_ET_test/" # Path to store figures/results
🏁 Initiate CATHY object
Read the DEM input file
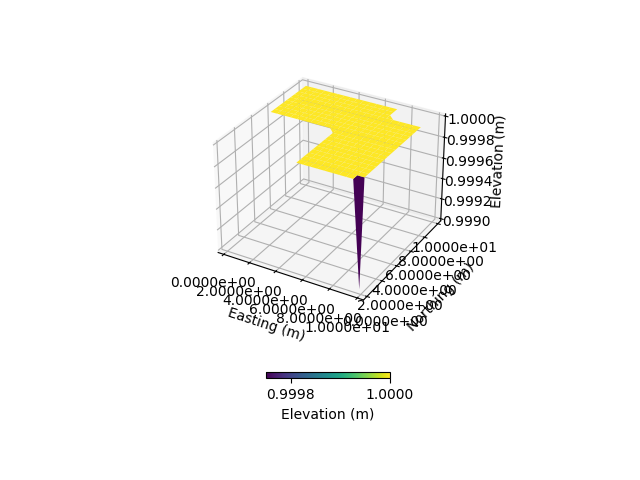
DEM, dem_header = simu.read_inputs('dem')
# Create a new DEM array filled with ones and add irregular boundary and invalid values (-9999)
DEM_new = np.ones(np.shape(DEM)) # Initialize new DEM with ones
DEM_new[-1, -1] = 1 - 1e-3 # Adjust a specific corner value
DEM_new[10:20, 0:10] = -9999 # Add an interior block of invalid values to simulate an irregular boundary
DEM_new[0:3, 15:20] = -9999 # Add an interior block of invalid values to simulate an irregular boundary
# Update the CATHY inputs with the modified DEM
simu.update_prepo_inputs(DEM_new)
# Visualize the updated DEM
simu.show_input('dem')
🔄 Update hap.in file
🔄 Update dem_parameters file
🔄 Update dtm_13 file
🔄 update zone file
🔄 Update dem_parameters file
🔄 Update parm file
🔄 Update dem_parameters file
Run the preprocessor to handle inputs and generate the mesh
simu.run_preprocessor()
# Create a 3D mesh visualization (VTK format)
simu.create_mesh_vtk(verbose=True)
# Load the 3D grid output
grid3d = simu.read_outputs('grid3d')
# Set parameters for elevation
simu.dem_parameters
elevation_increment = 0.5 / 21 # Define elevation increment per row
elevation_matrix = np.ones([21, 21]) # Initialize the elevation matrix
# Populate elevation_matrix with incremental values based on row index
for row in range(21):
elevation_matrix[row, :] += row * elevation_increment
🍳 gfortran compilation
👟 Run preprocessor
🍳 gfortran compilation
👟 Run preprocessor
wbb...
searching the dtm_13.val input file...
assigned nodata value = -9999.0000000000000
number of processed cells = 285
...wbb completed
rn...
csort I...
...completed
depit...
dem modifications = 281
dem modifications = 275
dem modifications = 269
dem modifications = 258
dem modifications = 245
dem modifications = 229
dem modifications = 208
dem modifications = 187
dem modifications = 173
dem modifications = 158
dem modifications = 144
dem modifications = 124
dem modifications = 101
dem modifications = 79
dem modifications = 56
dem modifications = 21
dem modifications = 0
dem modifications = 2808 (total)
...completed
csort II...
...completed
cca...
contour curvature threshold value = 9.99999996E+11
...completed
smean...
mean (min,max) facet slope = 0.000019190 ( 0.000000092, 0.002000000)
...completed
dsf...
the drainage direction of the outlet cell ( 7 ) is used
...completed
hg...
...completed
saving the data in the basin_b/basin_i files...
...rn completed
mrbb...
Select the header type:
0) None
1) ESRI ascii file
2) GRASS ascii file
(Ctrl C to exit)
->
Select the nodata value:
(Ctrl C to exit)
->
Select the pointer system:
1) HAP system
2) Arc/Gis system
(Ctrl C to exit)
-> ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dem file
min value = 0.999000E+00
max value = 0.100000E+01
number of cells = 285
mean value = 0.999997E+00
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
lakes_map file
min value = 0
max value = 0
number of cells = 285
mean value = 0.000000
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
zone file
min value = 1
max value = 1
number of cells = 285
mean value = 1.000000
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_w_1 file
min value = 0.000000E+00
max value = 0.100000E+01
number of cells = 285
mean value = 0.929825E+00
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_w_2 file
min value = 0.000000E+00
max value = 0.100000E+01
number of cells = 285
mean value = 0.701754E-01
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_p_outflow_1 file
min value = 0
max value = 8
number of cells = 285
mean value = 5.515790
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_p_outflow_2 file
min value = 1
max value = 9
number of cells = 285
mean value = 7.252632
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
A_inflow file
min value = 0.000000000000E+00
max value = 0.710000000000E+02
number of cells = 285
mean value = 0.299122810364E+01
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_local_slope_1 file
min value = 0.000000E+00
max value = 0.200000E-02
number of cells = 285
mean value = 0.141551E-04
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_local_slope_2 file
min value = 0.000000E+00
max value = 0.141421E-02
number of cells = 285
mean value = 0.998785E-05
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_epl_1 file
min value = 0.000000E+00
max value = 0.500000E+00
number of cells = 285
mean value = 0.492982E+00
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_epl_2 file
min value = 0.000000E+00
max value = 0.707107E+00
number of cells = 285
mean value = 0.528469E+00
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_kSs1_sf_1 file
min value = 0.000000E+00
max value = 0.240040E+02
number of cells = 285
mean value = 0.223195E+02
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_kSs1_sf_2 file
min value = 0.000000E+00
max value = 0.240040E+02
number of cells = 285
mean value = 0.168449E+01
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_Ws1_sf file
min value = 0.000000E+00
max value = 0.100000E+01
number of cells = 285
mean value = 0.929825E+00
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_Ws1_sf_2 file
min value = 0.000000E+00
max value = 0.100000E+01
number of cells = 285
mean value = 0.701754E-01
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_b1_sf file
min value = 0.000000E+00
max value = 0.000000E+00
number of cells = 285
mean value = 0.000000E+00
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_y1_sf file
min value = 0.000000E+00
max value = 0.000000E+00
number of cells = 285
mean value = 0.000000E+00
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_hcID file
min value = 0
max value = 0
number of cells = 285
mean value = 0.000000
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_q_output file
min value = 0
max value = 0
number of cells = 285
mean value = 0.000000
writing the output file...
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
dtm_nrc file
min value = 0.100000E+01
max value = 0.100000E+01
number of cells = 285
mean value = 0.100000E+01
writing the output file...
...mrbb completed
bb2shp...
writing file river_net.shp
Note: The following floating-point exceptions are signalling:
IEEE_UNDERFLOW_FLAG IEEE_DENORMAL
🔄 Update parm file
🛠 Recompile src files [12s]
🍳 gfortran compilation [18s]
b'/usr/bin/ld: cannot find -llapack: No such file or directory\n/usr/bin/ld:
cannot find -lblas: No such file or directory\ncollect2: error: ld returned 1
exit status\n'
😔 Cannot find the new processsor
👟 Run processor
b''
# Set up time intervals and cycles for the boundary condition
interval = 5 # Number of intervals
ncycles = 7 # Number of cycles
t_atmbc = np.linspace(1e-3, 36e3 * ncycles, interval * ncycles) # Time vector
# Atmospheric boundary condition value
v_atmbc_value = -2e-7 # Set the boundary condition value
# Check if the number of nodes matches the flattened elevation matrix
if int(grid3d['nnod']) == len(np.ravel(elevation_matrix)):
# Calculate the atmospheric boundary condition for each node based on elevation
v_atmbc = np.ones(int(grid3d['nnod'])) * v_atmbc_value * np.ravel(elevation_matrix)
else:
# For cases where the number of nodes doesn't match, calculate for all nodes
v_atmbc_all_nodes = np.ones(len(np.ravel(elevation_matrix))) * v_atmbc_value * np.ravel(np.exp(elevation_matrix**2))
# Reshape the boundary condition values to match the DEM shape
v_atmbc_mat = np.reshape(v_atmbc_all_nodes, [np.shape(simu.DEM)[0] + 1, np.shape(simu.DEM)[0] + 1])
# Mask invalid values in the DEM (-9999) by setting them to NaN
maskDEM_novalid = np.where(DEM_new == -9999)
v_atmbc_mat[maskDEM_novalid] = np.nan
# Flatten the masked matrix and remove NaN values
v_atmbc = np.ravel(v_atmbc_mat)
v_atmbc = v_atmbc[~np.isnan(v_atmbc)] # Use ~np.isnan to filter out NaN values
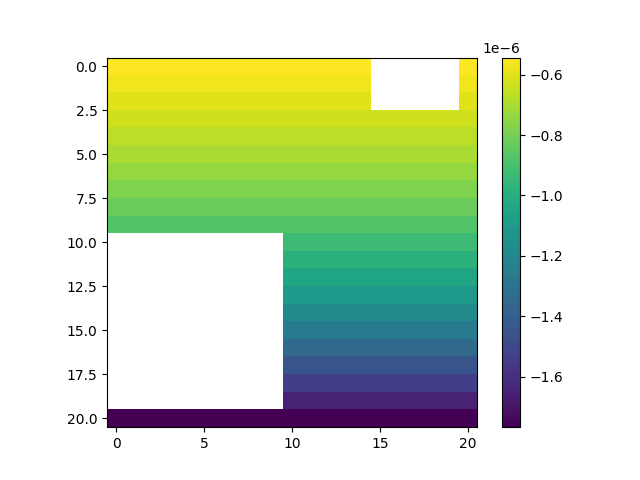
# Visualize the spatial variation of the atmospheric boundary condition
fig, ax = plt.subplots()
img = ax.imshow(v_atmbc_mat)
plt.colorbar(img)
# Update the atmospheric boundary condition (ATMB) parameters in CATHY
simu.update_atmbc(
HSPATM=0,
IETO=0,
time=t_atmbc,
netValue=[v_atmbc] * len(t_atmbc) # Apply the same boundary condition at all times
)
# Update the model parameters (time control) in CATHY
simu.update_parm(TIMPRTi=t_atmbc)
🔄 Update atmbc
🔄 Update parm file
🔄 Update parm file
────────────────────────── ⚠ warning messages above ⚠ ──────────────────────────
['Adjusting NPRT with respect to time of interests requested\n']
────────────────────────────────────────────────────────────────────────────────
# Run the model processor with specified parameters for time stepping and output control
simu.run_processor(
IPRT1=2, # Print results at time step 2
DTMIN=1e-2, # Minimum time step
DTMAX=1e2, # Maximum time step
DELTAT=5, # Time increment
TRAFLAG=0, # Transport flag off
VTKF=2, # Output VTK format
verbose=True # Turn off verbose mode
)
🔄 Update parm file
🛠 Recompile src files [18s]
🍳 gfortran compilation [24s]
b'/usr/bin/ld: cannot find -llapack: No such file or directory\n/usr/bin/ld:
cannot find -lblas: No such file or directory\ncollect2: error: ld returned 1
exit status\n'
😔 Cannot find the new processsor
👟 Run processor
b''
# Visualize the atmospheric boundary conditions in space using vtk
cplt.show_vtk(
unit="pressure",
timeStep=1, # Time step to display
notebook=False,
path=simu.workdir + "/atmbc_spatially_from_weill/vtk/", # Path to VTK files
savefig=True, # Save the figure
)
Traceback (most recent call last):
File "/home/runner/work/pycathy_wrapper/pycathy_wrapper/examples/SSHydro/plot_3d_spatial_atmbc_from_weill.py", line 136, in <module>
cplt.show_vtk(
File "/home/runner/work/pycathy_wrapper/pycathy_wrapper/pyCATHY/plotters/cathy_plots.py", line 557, in show_vtk
mesh = pv.read(os.path.join(path, filename))
File "/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/pyvista/_deprecate_positional_args.py", line 245, in inner_f
return f(*args, **kwargs)
File "/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/pyvista/core/utilities/fileio.py", line 267, in read
raise FileNotFoundError(msg)
FileNotFoundError: File (/home/runner/work/pycathy_wrapper/pycathy_wrapper/examples/SSHydro/atmbc_spatially_from_weill/vtk/101.vtk) not found
# Create a time-lapse visualization of pressure distribution over time
# cplt.show_vtk_TL(
# unit="saturation",
# notebook=False,
# path=simu.workdir + simu.project_name + "/vtk/", # Path to VTK files
# show=False, # Disable showing the plot
# x_units='days', # Time units
# clim=[0.55, 0.70], # Color limits for pressure values
# savefig=True, # Save the figure
# )
Total running time of the script: (0 minutes 24.663 seconds)